and Natural Sciences
Deep dive into plant signalling data questions potential of mobile mRNAs in plants
17 April 2025, by MIN-Dekanat
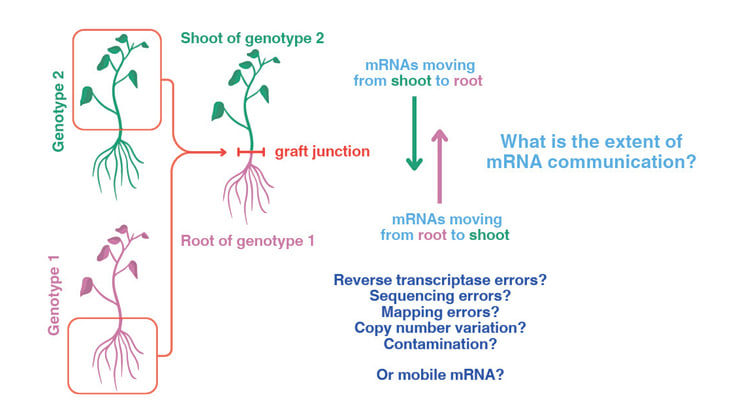
Photo: John Innes Centre
A far-reaching study with participation of the Department of Biology of University of Hamburg, has cast doubt on statistical methods used to identify long distance signalling networks in plants. The findings, published in Nature Plants, mean we may need to reevaluate a vital area of the life sciences.
The movement of messenger RNAs (mobile mRNAs) between cells and tissues has been reported to play a significant role in long-distance signalling in plants. Thousands of mobile mRNAs have been reported and proposed to act like a plant internet, as components of a complex intercellular communications system that regulates plant development and physiological processes. Understanding how plants communicate and respond to their environment has potential applications for agriculture, biotechnology, and even our fundamental understanding of evolution.
To determine the presence of these messaging molecules, researchers use a technique that involves grafting one plant onto another and tracking the progress of mRNAs as they pass from root to shoot, or vice versa, across the graft junction. The actual tracking is enabled by an experimental technique called RNA sequencing (RNA seq) which can identify differences in the sequences of mRNAs that might indicate the presence of mobile mRNA. However, a problem when analysing RNA seq data sets lies in distinguishing between differences in the sequences, which might arise from a graft-mobile mRNA signal, and “noise.”
To investigate, the team, led by the John Innes Centre (Norwich, England), used bioinformatics, genomic tools and machine learning approaches, devising a series of statistical tests to distinguish potential signals from noise in the data. In a study, meta-analysis of RNA seq datasets found that up to 83% of data obtained from grafted plants are indistinguishable from the sort of background noise that can be expected in this approach. Taking genome mis-mapping and potential contamination into account reduces the number of mobile candidates even further.
The finding that currently annotated mobile mRNAs lack statistical basis challenges current thinking on the extent of mRNA communication. Professor Richard Morris, corresponding author of the study said: “Our analysis questions the number and signaling potential of mobile mRNAs in plants.”
The aim was to develop models for mRNA transport but the lack of features in the data motivated the researchers to take a closer look. But the deeper they dug, the less support they found for mobile mRNA. Following these findings, the team have put together a set of recommendations to help future researchers develop more robust approaches for studying long distance mRNA transport.
The paper is a major result of the European Research Council Synergy project PLAMORF (Plant Mobile RNAs: Function, Transport and Features) and all three Principal Investigators - Julia Kehr, Richard Morris and Friedrich Kragler - are co-authors of the study.
Text: John Innes Centre, red.


