and Natural Sciences
DNA Origami Signposts
31 March 2021, by MIN-Dekanat
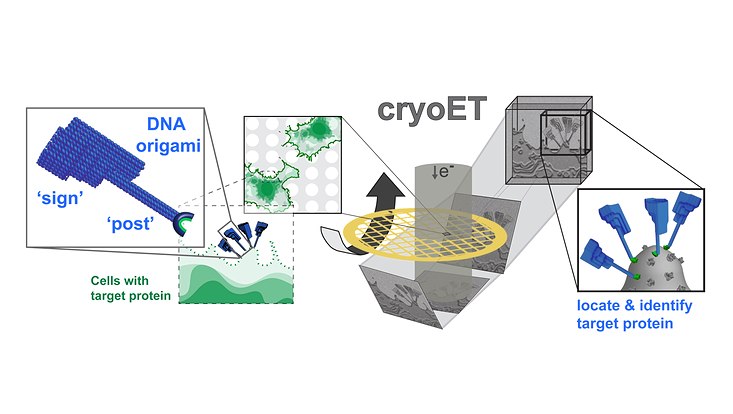
Photo: Modified from Silvester et al, 2021
Researchers from Centre for Structural Systems Biology’s Grünewald group (Heinrich Pette Institute / Universität Hamburg) in collaboration with researchers at the University of Oxford, recently developed a method to pinpoint individual protein structures using DNA origami nanostructures and electron cryotomography (cryo-ET).
Cryo-ET reveals the native molecular interactions that take place in crowded biological environments such as membranes, cells and viruses. “However, the noisy nature of the data makes it difficult to identify small features of interest such as individual proteins.” explains Kay Grünewald.
Working with collaborators in the Turberfield group in the Department of Physics, University of Oxford, experts in DNA origami, the researchers designed a signpost-shaped DNA-nanostructure. “The ‘sign’ provides a distinctive signal that is easily detected in cryo-ET data,” explains Lindsay Baker (University of Oxford/ HPI), “while the bottom of the post specifically recognizes commonly used fluorescent protein tags.”.
“Many important biological interactions take place on cellular surfaces.” says Grünewald. “We now have a reliable cryo-ET method to help identify the specific proteins initiating and driving these interactions; and without the need to know their structure or conformation a priori.”
Text: CSSB
Original publication
Silvester E, Vollmer B, Pražák V, Vasishtan D, Machala EA, Whittle C, Black S, Bath J, Turberfield AJ, Grünewald K, Baker LA (2021)DNA origami signposts for identifying proteins on cell membranes by electron cryotomography. Cell.184(4):1110-1121.e16.


